NWB Format#

Platforms |
Windows, Linux, macOS |
Built in? |
No |
Key Developers |
Aarón Cuevas López, Pavel Kulik, Josh Siegle |
Source Code |
Advantages
NWB is a widely used format for sharing data among neuroscience labs.
Data is stored in a single HDF5 file with self-documenting internal structure.
Files can be read using the pynwb or matnwb libraries, or with the growing number of high-level tools that support the NWB format.
Limitations
HDF5 files must be closed gracefully, so data may be irrecoverable if the GUI crashes during acquisition.
The HDF5 C++ library is not thread-safe, so you cannot write to the NWB format from multiple Record Nodes simultaneously.
File organization#
Within a Record Node directory, data for each experiment (stop/start acquisition) is contained in a separate NWB file. Individual recordings are appended to datasets stored inside the “acquisition” group.
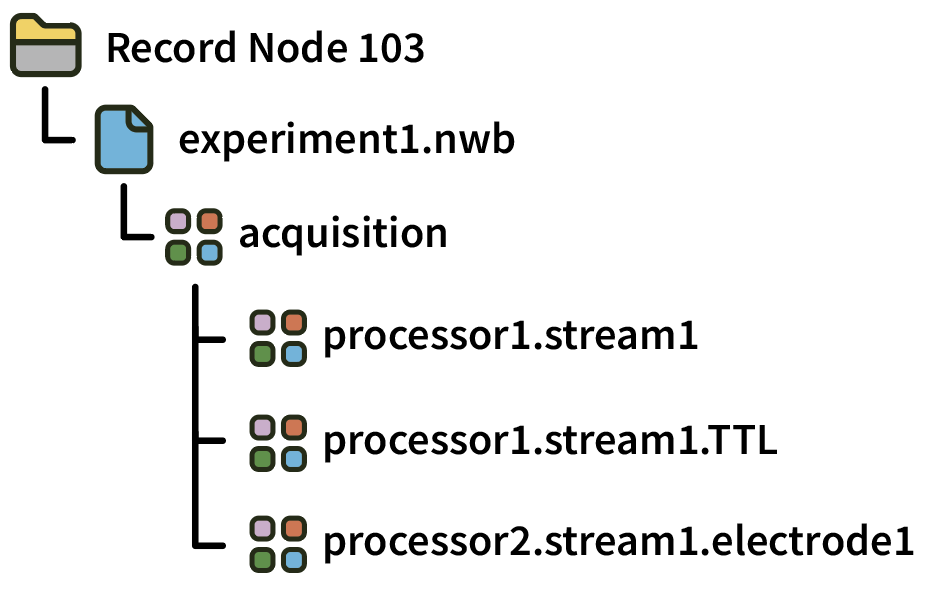
Each NWB file also contains the following information:
/file_create_date: date + time in ISO format (text array)/identifier: string identifier for this file (text array)/nwb_version: ‘2.4.0’ (text attribute)/session_start_time: date + time in ISO format (text array)
Format details#
Continuous#
Continuous data is grouped by stream (a block of synchronously sampled channels):

Each continuous group is an NWB ElectricalSeries containing the following datasets:
data: N channels x M samples of 16-bit integers. Thechannel_conversiondataset stores the “bitVolts” value required to convert these values into volts.timestamps: M 64-bit floats representing the timestamps (in seconds) for each sample.
Events#
Event data is organized by stream and event channel (eusually named TTL). Each event channel can contain data for multiple TTL lines.

Each events group is an NWB TimeSeries containing the following datasets:
timestamps: N 64-bit float representing the timestamps (in seconds) for each eventdata: N event codes indicating ON (+CH_number) and OFF (-CH_number) statesfull_words: N 64-bit integers representing the state of the first 64 TTL lines when each event occurred.
Spikes#
Spike data is organized by stream and electrode.

Each spikes group is an NWB SpikeEventSeries containing the following datasets:
data: array with dimensions S spikes x N channels x M samples containing the spike waveforms. Thechannel_conversionattribute stores the “bitVolts” value required to convert these values into microvolts (headstage channels) or volts (ADC channels).timestamps: S 64-bit floats containing the timestamps (in seconds) corresponding to the peak time of each spike.
Reading data in Python#
Create a
Sessionobject using the open-ephys-python-tools package. The data format will be automatically detected.
Reading data in Matlab#
Use the open-ephys-matlab-tools <open-ephys/open-ephys-matlab-tools>`__ library.
Note
NWB files written by the Open Ephys GUI are not currently compatible with the MatNWB library. We are working on a fix!